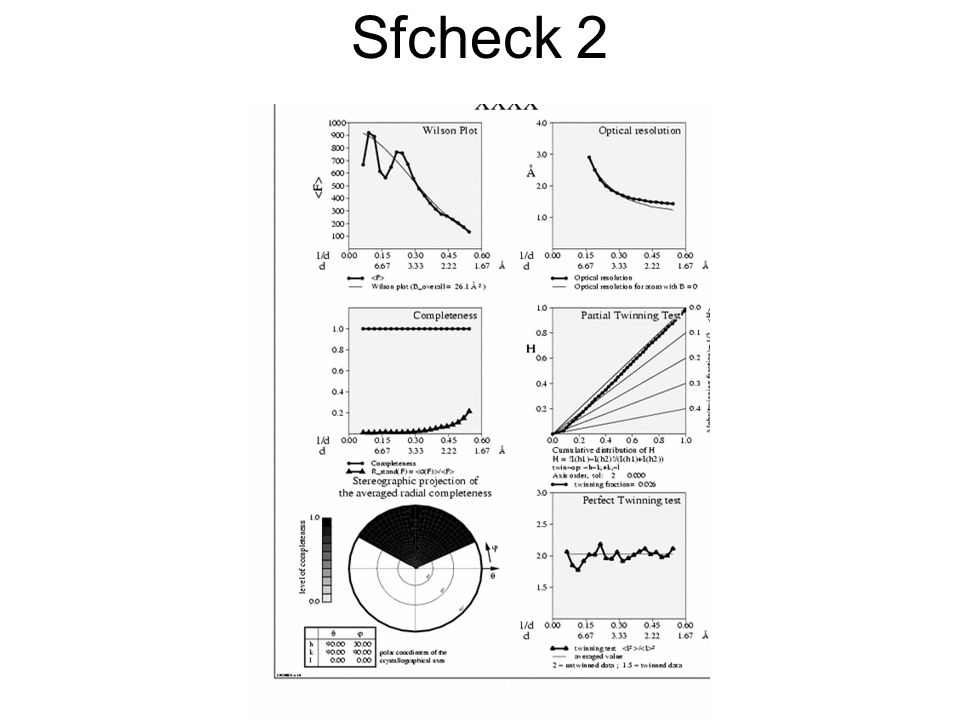
Plotting the optical resolution against the nominal resolution upper right-hand plot in Fig. Removing these data entirely from the calculations produces series-termination effects, which introduce spurious peaks in the electron density at the surface of the macromolecule. Inspection of the corresponding region of the electron-density maps, displayed in Fig. The local agreement between the model and the electron-density map is evaluated on a per-residue basis, considering separately the macromolecule backbone and side-chain atoms, as well as solvent atoms and heterogroups. All members of the European Consortium on Structure Validation are thanked for stimulating discussions. In the following, we provide a detailed account of the structure-factor data analysis and the scaling procedures, and describe the different parameters and procedures used by SFCHECK in assessing the quality of the model as a whole and in specific regions. Small values of D corr , depicted by tall bars in Fig.
| Uploader: | Akinora |
| Date Added: | 18 February 2016 |
| File Size: | 8.96 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 30898 |
| Price: | Free* [*Free Regsitration Required] |
B factor and connect chain connectivitycomputed considering the regular electron densities see legend of Figs.
Why is there a difference between the Rfactors from Refmac and Sfcheck? - CCP4 wiki
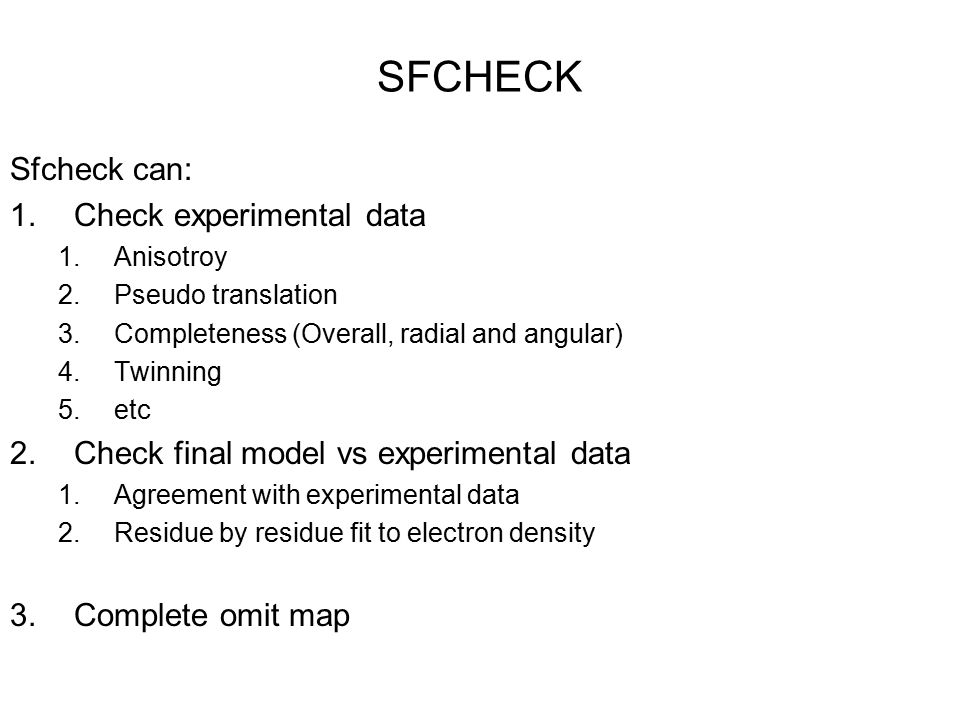
In this paper we present SFCHECKa stand-alone software package that features a unified set of procedures for evaluating the structure-factor data obtained from X-ray diffraction experiments and for assessing the agreement of the atomic coordinates with these data. R factor, R free and the correlation coefficient To evaluate the global agreement between the atomic coordinates and the electron-density map, SFCHECK computes three well known parameters commonly used as indicators for the quality of X-ray structures of macromolecules.
The meanings of the various listed quantities are either self-explanatory or described in the text. In addition, it gives several estimates of the average error in the atomic coordinates. The Interface will derive a name from the input MTZ filename, but you can change it. Comparison of the B -factor and density index plots can be useful for detecting regions with errors in the sfchfck.
Given the diversity of the latter formats, human intervention is often necessary to process these files correctly. Connect measures the level of the sfchek density along the macromolecule skeleton and can be used to assess the continuity of the electron density along the polymer chain.
SFCHECK (CCP4: Supported Program)
Nature London, — The latter task, in particular, is notoriously difficult, and the criteria proposed here by SFCHECK should be considered only as a starting point for more detailed analyses. The cutoff function varies between zero and one, in the manner shown, and equals 0.
For more information, click here. Prior permission is not required to reproduce short quotations, tables and figures from this article, provided the original authors and source are cited. Among the criteria are the normalized average atomic displacement, the local density correlation coefficient and the polymer chain connectivity.
The segment 98— also displays significantly higher backbone and side-chain B factors, as well as poorer backbone connectivity. Indeed, it allows the identification of several features which could be the sfchexk of difficulties in sfchec structure determination.
See the entry in the refinement module for further details. Plotting the optical resolution against the nominal resolution upper right-hand plot in Fig. The high R stand F values at resolutions higher than 3. The Determination of Crystal Structures. It then uses FFT to compute two electron-density maps, with calculated phases and observed and calculated amplitudes, and calculates fscheck gradients of the difference maps with respect to atomic coordinates.
The electron-density correlation coefficient, D corrfor a given group of atoms is calculated as. Several such procedures have been developed in recent years. The dependence of various parameters on the nominal resolution d spacing is also given. The same plot in Fig.
Proteins26— With the shape of the atomic peak being fitted by a single Gaussian, this minimum distance equals twice the standard deviation of the fitted Gaussian, or its width W. Expected maximal error Following the method of Cruickshankthe standard deviation of the atomic coordinates can be derived from the properties of the electron-density map.
Following the method of Cruickshankthe standard deviation of the atomic coordinates can be derived from the properties of the electron-density map.
Low levels of the connect index indicate locations where this continuity is broken. DPI, diffraction-data precision indicator DPI is the atomic coordinate error estimated by the method of Cruickshank.
To further assess the quality of the structure-factor data, the program computes five additional quantities: All columns will be output to the mmCIF file, but these can be flagged in a number of ways.
This particularly applies to the atomic coordinates of the macromolecules.

Комментариев нет:
Отправить комментарий